My Contributions to the Newt Project Part 2
This is the second part of my contributions blog post.
Introduction
This is the second part of my blog post logging the contributions I made to the Newt project. You can see that post here. This blog post will go mostly into the contributions I made after the work I outlined in the first blog even though there might be some overlap. Let’s go through it:
Small Bug Fixes
GitHub Issue There was a small bug related to loading maps on top of each other when they had certain color schemes. I found this bug and resolved it.
GitHub Issue There was a small bug related to a spinner (which is a small animation indicating a load screen) that appears if we try to apply layout an empty map. This was a nice and easy fix.
`Added at the time of the edit, 13th of August GitHub Issue There was a small issue with the SBGN bricks feature, which, as the name suggests, should only work with SBGN formats (specifically, PD). However, it worked with SBML also, which is not intended. I disabled this behaviour.
Build Box
Github Issue This was a suggestion by me. In web development, it is usual to have an internal server (meaning only the people working in a company have access to that server). This server occasionally gets rebuilt (which means updated with the latest changes). In our team, there was some confusion about when this server was last updated, what changes are currently on the internal server ready to be tested, etc… I suggested to include a build box that included the time of the last rebuild of the internal server. It looks like this:
as you can see, in the blue portion in the middle, we have the build information.
File Loading Improvements
GitHub Issue I made two suggestions to include in the next Newt release. First one is including the spinner when we are trying to import very large files.
The second one is related to the drag & drop import operation. This operation previously only worked for a limited number of file formats. For the other file formats, we had to use the import menu. I added functionality so that we don’t have to use the import menu and just use the drag & drop functionality. This issue was mostly resolved, but there were some concerns related to code duplication (which is when we write code that solves the same problem in different parts of the software). The remaining part of the issue was taken out of the release milestone, which just means it will be revisited in a later time.
SBML UI, Palette and Legend Improvements
GitHub Issue It was found that there were significant issues with the SBML (which is a file format) support in Newt. Couple of issues were opened about these problems, one of which is this one. This one is generally about visual enhancements for the users. It included six items to be fixed, to which I also added one. Resolving this issue was a relatively long process, which involved careful frontend work. After a lot of back and forth trying to understand and resolved the issues, we managed to completely fix this issue.
I also suggested a visual update of labeling the nodes / edges in the palette. Below, you can see the old (top) and the new (bottom) images
Import / Export Operation Improvements
GitHub Issue I was originally not going to do this, but I was included anyway. This is the largest standing issue on this project since I have started my work. A complicated issue that encompasses a wide variety of functionalities that need to be fixed.
Let me try to explain what this is. There are many file formats for doing systems biology. I went over this concept in my previous blog post. These formats can sometimes be converted to each other. In Newt, we have support for this type of functionality whenever it is possible. However, we don’t do all of this ourselves. Sometimes, we use external services to do our converting. The aim is getting a file which represents the model you have on the screen. This is broadly called exporting. Some issues were discovered with these processes. The reverse, loading an exported file into the app, is called importing. Some issues were also discovered with the import process.
The biggest issue here was that we were not exporting nor importing layout information (which is related to how we show a biochemical map to our users, what goes where etc…) when we are working with the SBML file format. I was included to fix the export side of this issue. In order to learn how these formats work (by extension learning how they might be exported), you need to read technical documents called specifications. Now, those of you that are not familiar to documentations and specifications might not be aware of this, but those documents are usually long and dense. This is the SBML Level 3 Version 2 Specifications, which is 180+ pages long. The good news is that we don’t have to read all of it. We try to only read what is relevant to us (which is usually not very straightforward to determine). There are also good software packages that help us in the process. Luckily, this was the case. Even though this issue is still ongoing at the time of writing (but almost resolved at the time of the edit, -UUE), we have made major progress. I have also upgraded our app to use SBML Level 3 Version 2, which is a more thorough standard for what we want to achieve (it is also the latest).
At the time of the edit, I was tasked with and resolved more items on this issue, which included a PD to SBML (and vice versa) conversion using the MINERVA service. I also implemented something called infer nesting for SBML, which is a graph algorithm to put smaller nodes inside bigger nodes.
Auxiliary Box / State Information in Export / Import
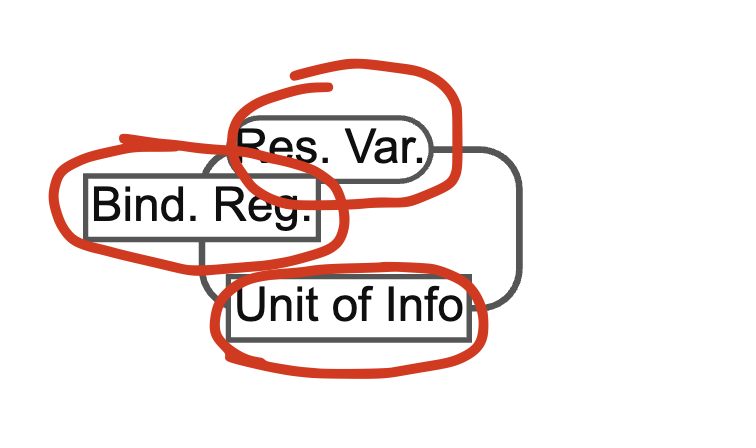
GitHub Issue, GitHub Issue I have already talked about SBML export / import, which is also what the focus of this issue is. In our interpretation of the SBML format, following CellDesigner, we have auxiliary boxes. See the marked regions in the image below.
These are the binding region, residue variable and the unit of information. There is also something called state information for nodes. See picture below:
these are the multimer node, the active node and the hypothetical node (in that order, left to right). These information were not exported previously. I was tasked to export and import them.
Now, there is a reason why I said that this was our interpretation of the SBML format. That’s because the SBML format is not exact with regards to what visual features we put on the screen. Visually representing a biochemical map is only a secondary concern with SBML. It is more concerned with the integrity of the underlying model which consists of species and reactions (broadly). The format is designed for a machine to understand and simulate whereas something like the SBGN format is designed not to simulate but to show the user what the biological model is.
Why have I explained all this? Well, I want to express that there is not an exact way of storing this visual information in the SBML format even though Newt has these SBML features, which is fine. As I said, it is not an exact standard with respect to the visual features. Luckily, the people who develop this SBML format are smart people and recognize that individual softwares might want to go above and beyond what the intention of a pure SBML file would be. They include a way of annotating elements through the <annotation> tag. The contents of this tag can freely ignored by other software packages, which means it includes non-essential software specific information about the underlying model. This is exactly what I am trying to do with this issue.
With all that out of the way, I started working on our annotation scheme. It has some rules outlined in that 180+ page document, but it is not very limiting at all (basically any valid XML with a namespace goes). I developed our format for this and implemented export / import functionality for our software. As of the writing of this blog, this issue is awaitig to be reviewed and closed (it is closed at the time of edit, -UUE).
Conclusion / Future Work
Well, this was a good amount of work that I outlined. When reading through the SBML documentation, I realized that the most important feature of the SBML format is producing simulations for biochemical reactions. However, there is currently no support for this in Newt. I talked to my mentor and we agreed to talk about this after the release because it would be a long process and delay the release unnecessarily.
At the time of edit, 13th of August, I have prepared a document for SBML simulation support and sent it to my mentor. I expect that the new Newt release will come sometime this month.
At the time of second edit, 20th of August, we decided to move forward with the simulation features as outlined in the document. I expect that I will be working to add that feature over the course of the semester, but we will see.
This blog also has more readers than I anticipated initially (Which is admittedly 0. I opened this blog mostly for documenting my work, though I am planning on expanding my scope to talk about books, mathematics or other stuff). I might add a mailing list for people who are interested in being notified when my blogs come out. I might also share an Instagram story about my blog. We will see. Definitely let me know if you are interested in a mailing list (If you are reading this, you can probably access me on either Instagram or Whatsapp).
Have a lovely rest of the summer!